Past Projects
Goals of our applied projects include management and prevention of spread of invasive species and disease. Second important area for our applied research is ecological genetics where we particularly focus on cryptic biodiversity.
We used the zebra mussel D. polymorpha as a model species to predict quagga invasion. The zebra mussel is a close relative of the quagga mussel and has invaded Switzerland several decades ago. By investigating its distribution, genetic structure and life history traits we revealed potential distribution vectors, barriers and limiting factors for the population establishment and growth of the quagga mussel. We compared the niche constraints of quagga and zebra mussels directly in experiments using mesocosms.
Predicting the Dreissena Invasion to Switzerland
The quagga mussel Dreissena bugensis is highly invasive in North America and Europe. It has already been found in the Upper Rhine at Karlsruhe and most likely, it will be invading Switzerland in the next few years. In other regions, where this mussel has already spread, it is known to have a strong negative impact on ecosystems, biodiversity and socioeconomics. Aim of our project is to provide the basic knowledge to predict the future distribution and impact of the quagga mussel in Switzerland.
Such data should allow to predict the future quagga mussel distribution. Our additional aim is to develop molecular tools for early detection of the quagga larvae in the water column, which is essential for providing an early warning system for authorities. Altogether the data will help to define monitoring and prevention strategies against the spread of the quagga mussel. With this case study we hope not only to derive guidelines for management of aquatic invasive species in Switzerland, but also to gain experience from the interaction with the key stakeholders who will need to take measures due to problems caused by aquatic invasive species.
Disciplines: invasion biology, population genetics, freshwater Ecology
Keywords: invasive species, Dreissena, distribution, spread, source-sink, life history traits
Responsible persons: Lukas De Ventura, Kirstin Kopp
Please aproach someone from our current members for contact details.
The Universe Inside
or: Microbiomes are everywhere
Host-microbiome interactions represent a crucial factor in shaping the ecology and evolution of the arthropods. Especially for detritivorous isopods, microbial symbionts help them to overcome the challenges posted by low-nutrient detritus diet by providing essential digestive enzymes. Here, we examining the role of microbiome in facilitating detritivorous diet and its interactions with host health.
Aijuan Liao (then MSc student) compared the microbiomes of hindguts and midguts of acanthocephalan infected and uninfected Asellus aquaticus.
Discipline: host-microbiome interactions
Keywords: Asellus aquaticus, microbioal symbionts
Responsible person: Claudia Buser
Ecology and Evolution of Pathogen Resistance in Changing Environments
Environmental factors such as habitat quality and environmental stress (e.g. reduced availability of external resources for hosts) have been suggested to play a major role in host–pathogen ecology and evolution, but the roles of several other factors such as anthropogenic climate change are poorly understood. Due to climate change, temporal variation in air temperatures is expected to increase so that extreme weather conditions such as heat waves become more frequent. This can induce a strong selective pressure on natural populations and affect host–pathogen interactions. In this project, we will examine how exposure to high temperatures affects the maintenance of host immune defences, and how alterations in immune functions are connected to other life history traits (e.g. reproduction, longevity). Furthermore, we will investigate if host populations show within and between population genetic variation in thermal tolerance, and if they thus could be able to adapt to changing environmental conditions. Proposed studies will use a freshwater snail (Lymnaea stagnalis) as a natural model system. This system allows quantitative genetic studies and detailed immunological work. Our approach in this project is experimental; by manipulating the ambient temperature, we create environments where the degree of host stress varies. Proposed project is tightly connected to the larger research protocol of our collaborative network (collaborators in Switzerland and in Finland), which enables excellent facilities and research environment for this project.
The common goal of the proposed project is to understand the interplay between environmental and genetic factors determining ecological and evolutionary processes in host–pathogen relationships. Understanding these processes can have a key role in the combat against disease outbreaks in natural populations as well as in industry.
Disciplines: ecology, evolution
Keywords: parasite-host interactions, immunoecology, temperature, freshwater snails
Contact person: DownloadOtto Seppälävertical_align_bottom
Ecological genetics of Bryozoa-Myxozoa host-parasite interactions
Understanding the clonal and population genetic structure of the freshwater bryozoan Fredericella sultana is decisive for understanding the epidemiology and management of the myxozoan parasite Tetracapsuloides bryosalmonae that causes the Proliferative Kidney Disease (PKD) in wild and farmed salmonids. T. bryosalmonae infections in F. sultana cause reductions in growth and reproduction, but rarely lead to mortality. In rainbow trout the parasite causes high mortalities in temperatures above 15˚C and impacts wild trout populations across Europe. The interaction between F. sultana and T. bryosalmonae is pivotal for the spread, prevalence and incidence of the disease in fish. F. sultana is a sessile, filter-feeding, colonial invertebrate, with a largely clonal reproductive strategy. Common, widespread and susceptible F. sultana clones may serve as a persistent parasite reservoir, supplying disease epidemics in fish, yet, at present, no information of the clonal structure of F. sultana populations exists. To predict the impact of environmental change on spread of PKD in fish, it would be essential to resolve questions regarding the geographic distribution of susceptible and resistant F. sultana genotypes, and their potential response to environmental change. For prediction of co-evolutionary responses, e.g. evolution of resistance in F. sultana, estimates of the magnitude of gene flow among the F. sultana populations are also necessary. At present such information is not available due to lack of neutral genetic markers for F. sultana. We are (i) developing the required nuclear genetic markers that will allow high resolution genetic studies of the clonal and population genetic structure of F. sultana populations, and (ii) use these markers in the first genetic surveys of F. sultana populations that are either known to be infected by T. bryosalmonae or known to be infection-free, based on earlier ecological surveys. Results of the study will be highly beneficial for preventive and predictive applications to manage PKD in salmonids.

Disciplines: population genetics, fish diseases, river ecology, conservation biology
Keywords: ecology of disease, PKD, trout, bryozoa, myxozoa, clonal structure
Contact person: DownloadHanna Hartikainenvertical_align_bottom
Ecological Genetics and Adaptation
We are interested in how the (population) genetic structure is influenced by the environment or the interaction with organisms.
Phenotypic Selection and Quantitative Evolutionary Responses in Immune Defence
Immune defence is considered as an important trait for organisms’ fitness as it eliminates harmful parasites. Therefore, a strong immune defence is often assumed to evolve as a response to parasitism. However, since immune defence is energetically costly to maintain and use, immune defence could be under stabilising selection. Although understanding the type and strength of natural selection on immune defence traits is in high demand for predicting their evolutionary responses, this information is still generally lacking in most natural systems. In this project, phenotypic selection and its relation to quantitative genetic variation in host immune defence traits (i.e. their expected evolutionary responses) is studied in natural populations of a freshwater snail Lymnaea stagnalis. The research concentrates on novel aspects of evolution of immune defence by examining, for example, the role of variation in infection risk and amount of genetic variation (estimated using neutral markers) among host populations, and different parasite types in these processes.
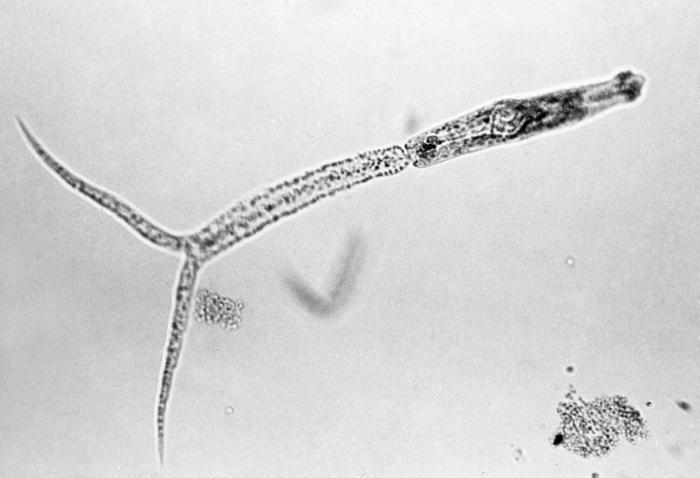
Disciplines: ecology, evolutionary biology, ecological immunology
Keywords: host-parasite interactions, life history evolution, natural selection, fitness, immune defences, freshwater snails
Contact person: DownloadOtto Seppälävertical_align_bottom